AbstractSleep is a crucial and evolutionarily conserved phenomenon, but the mechanisms that control sleep-wake behavior and underlie sleep disorders are not yet fully understood. One major challenge for sleep research was the lack of technology that allows for cell-type- and circuit-specific investigation of neurons and neural circuitry. A decade ago, a novel methodology known as optogenetics was developed, which uses light to control specific cell types of neurons, either to activate or inhibit neuronal firings. The strength of optogenetics in neuroscience is the precise control of neuronal activities in millisecond scale and the ability to dissect the neural circuits and cell types to understand their functions. There have been substantial advancements made in the field of sleep research through the implementation of optogenetics. This review provides a brief introduction on the optogenetics and a consolidated summary of recent findings published in sleep research using optogenetics.
INTRODUCTIONSleep occupies more of a person’s time than any other single activity with the average human spending nearly one-third of his or her life asleep [1,2]. Sleep plays an essential role in maintaining good mental and physical health, yet sleep disorders are currently among the most prevalent clinical problems [3]. Over the past several decades, several breakthroughs have led the deeper understanding of sleep and sleep medicine. Recent findings reveal that at least eight hours of sleep is advantageous for behavioral alertness and working memory, whereas sleep deprivation contributes to declines in memory and attention [4]. Research on sleep loss in Australia calculated an economic burden exceeding 5 billion dollars through healthcare expenses, loss of productivity costs and other factors associated with sleep disorders [5]. Thus, any progress towards better understanding the cellular and molecular systems involved in sleep regulation could have a potential to be translated into clinical benefits.
Recent advances have been made investigating the cellular mechanisms and neural circuitry implicated in sleep functions, largely attributed to the increased implementation of optogenetics, which is emphasized in research targeting specific neuronal populations [3]. In addition to sleep research, optogenetics, the utilization of optics to activate photosensitive proteins expressed in target cells thereby activating or inhibiting neuronal firings, has been applied in studies of γ-aminobutyric acid (GABA)-ergic neurons and cholinergic neurons of the basal forebrain (BF), hypocretin (Hcrt)-expressing neurons of the lateral hypothalamus (LH), serotoninergic dorsal raphe nuclei (DRN), noradrenergic locus coeruleus (LC), and a variety of other cells implicated in the regulation of wakefulness and arousal (Fig. 1) [6].
Deciphering the relationships between separate populations of neurons is a critical component to determining the roles specific neuronal subsets play in sleep, as well as their potential uses in sleep medicine. This review aims to discuss recent publications in the field of sleep research involving optogenetic techniques. We seek to provide a summary of recent sleep studies, exploring the future direction of sleep research and addressing some of the current challenges.
A BRIEF OVERVIEW OF OPTOGENETICSOptogenetics brings forth a new era of technology enabling heightened precision with regards to temporal and spatial control in the activation (depolarization) or inhibition (hyperpolarization) of specific groups of targeted neurons. It has been applied widely in the field of neuroscience for the mapping of neurocircuitry and investigation into the roles of key neurons in certain neuropathologies. Two major advantages of optogenetics are the millisecond scale precise control of neuronal activities using opsins and light, and the cell-type specific controllability using molecular and genetic techniques.
Precise Control of Neuronal Activities using Opsins and LightThe opsin is a protein which is sensitive to the light exposure and has the ability to generate biological responses, such as opening of membrane channels or activation of second messenger system. Currently, there are a wide range of natural and genetically engineered opsins available for a variety of purposes. There are two major kinds of opsins which are being used in optogenetics: activating or silencing opsins. First, channelrhodopsins (ChR), light-gated nonspecific cation channels, are activating opsins that can cause the influx of positively charged ions, such as sodium ion, into the neurons through the opsin pores when exposed to 473 nm blue light, resulting in an excitatory depolarizing effect [7]. Researchers have been able to generate several ChR variants to achieve specific purposes, including ChR2 for the first experiment to excite neuronal membrane with light [8], step function opsin (SFO) for the bi-stable excitation [9], stabilized SFO for the longer channel opening time, ChETA for ultrafast optogenetic control, and C1V1 composed of ChR1 and VChR1 fragments to enable red-shifted optical excitation. Second, halorhodopsins (NpHR) and archaerhodopsins (Arch) are silencing opsins. NpHR pumps chloride ions into neurons and Arch pumps the protons out of neurons, resulting in the decreased intracellular membrane potential (hyperpolarization) and the neuronal silencing or inhibition. Initially, NpHR was challenged with issues concerning phototoxicity [10] and cellular localization [11]. These concerns were addressed in later variants of NpHR including enhanced NpHR2.0 and 3.0 (eNpHR2.0 and eNpHR3.0), which were engineered for enhanced membrane targeting, larger photocurrents and to generate lower phototoxicity [10]. Arch from Halorubrum strain TP009, also referred to as ArchT, are also proton pumps that have high inhibition capacity [11]. Within this past decade, high-resolution crystal structure imaging systems enabled structure-guided engineering of ChR2 and the generation of chloride-conducting ChR.
Cell-Type Specific ControllabilityAnother advantage of optogenetic technique is the ability to control certain types of neurons leaving other neuronal types unaffected [12,13]. To perform cell-type specific optogenetic control, opsin should be expressed in only specific types of neurons. Viral vectors can be used to deliver exogenous genes into the cells. In optogenetics, viral vectors that contain the promoter of a marker protein gene, e.g., choline acetyltransferase for cholinergic neurons, and opsin gene are stereotaxically injected to a target area of the brain. Several types of viruses are being used, including adeno-associated virus, lentivirus, herpes viruses and rabies viruses. The other strategy is to use the Cre-dependent expression system [14]. Animals expressing Cre recombinase in a specific cell type can be used for this approach. Viral vectors with double-floxed inverted opsin gene make the opsin expressed only in the presence of Cre recombinase. Consequently, opsins can be expressed in a specific cell type. Lastly, transgenic mouse lines that express opsins in specific cell types can be used [15]. These mouse lines are genetically engineered and born with the opsin expressed in the targeted cell type.
NEURAL CIRCUITRY ASSOCIATED WITH SLEEP-WAKE CONTROLSleep-wake behaviors are controlled by interactions between multiple regions of the brain [16]. Cell groups located in the brainstem, BF, thalamus and hypothalamus are critical for stimulating cerebral cortex and generate non-rapid eye movement (NREM) and rapid eye movement (REM) sleep states [17]. Originating at the rostral pons, the ascending arousal pathway contains two major branches [17]. The first branch involves pedunculopontine and laterodorsal tegmental nuclei cells (PPT and LDT, respectively), which provide input for activation of the thalamic relay neurons required for transmitting information to the cerebral cortex [17]. Rather than using the thalamus, the second branch activates BF and LH neurons [17]. This branch begins with a subset of monoaminergic neurons of the upper brainstem and caudal hypothalamus, including histaminergic (HA) tuberomammillary nucleus (TMN), dopaminergic ventral periaqueductal gray matter, serotonergic dorsal and median raphe nuclei, noradrenergic LC and more [17].
In contrast, the ventrolateral preoptic nucleus (VLPO) is associated with promoting sleep [17]. In the 1990s, researchers discovered that the VLPO was one of many monoaminergic cell groups responsible for neuronal signaling patterns connected to sleep [17]. Neurons in the VLPO project to major neuronal clusters in the brainstem and hypothalamus that are associated with arousal and inhibit the arousal-related nuclei by releasing inhibitory neurotransmitters such as GABA and the neuropeptide galanin [17–19]. It is possible that damage to the VLPO has the potential to result in insomnia [17]. VLPO neurons can be inhibited by serotonin and noradrenaline [20]. The seemingly abrupt transitions between sleep and wakefulness may be caused by the “flip-flop” circuit involving mutually inhibitory components of the sleep-wake system [17]. In wakefulness, the VLPO is inhibited by the monoaminergic nuclei, resulting in disinhibition of orexin neurons, LDT/PPT, and the monoaminergic neurons themselves [17]. In sleep, VLPO actively inhibits monoaminergic cells and orexin neurons, which together decrease the likelihood of sleep interruptions [17]. The sudden and complete transitions from a state of sleep to wakefulness prevents the potential dangers associated with impaired alertness or inefficiency of half-awake sleeping periods [17].
WAKE-RELATED NEURONS DISCOVERED WITH OPTOGENETICSAfter optogenetic methodology was introduced in sleep research, studies on sleep-wake transitions have revealed that a number of brain regions and their various cell types are associated with promoting or maintaining wakefulness, such as Hcrt or orexin neurons of the LH, noradrenergic neurons of LC, the serotonergic or dopaminergic neurons of the DRN [21,22], HA and GABAergic neurons of the TMN [23], cholinergic, glutamatergic and parvalbumin (PV)-containing neurons of the BF [24–26], and dopaminergic neurons of the ventral tegmental area (VTA) (Table 1) [27]. The circuitry associated with wakefulness is very complex and often involves a variety of pathways.
To determine whether a neuron group is associated with wakefulness, researchers can optogenetically stimulate neurons of interest while a subject is under a sleep state in order to identify if the neurons can cause sleep-to-wake transition. A second strategy is to inhibit a group of neurons while a test subject is awake and determine if it induces a sleep state. The former is a gain of function test which can prove whether the activity of target cells is a sufficient condition for the wakefulness, whereas the latter is a loss of function test which can verify whether the activity of the target cells is a necessary condition for the wakefulness.
Hcrt-expressing neurons and noradrenergic LC neurons are two neuronal populations of great interest in optogenetic sleep research. Hcrts, also called orexins, are hypothalamus-specific peptides with neuroexcitatory activity, which play a critical role in maintaining wakefulness [28,29]. In 2007, Adamantidis et al. [30] demonstrated the first in vivo application of optogenetics in sleep research in which they made ChR2 expressed only in ChR2 Hcrt neurons of LH and optically stimulated ChR2 to investigate the wake-promoting effect. They showed a causal link between the activation of Hcrt-expressing neurons and transitions from NREM and REM sleep to wakefulness [30]. The wake-promoting effect was maintained throughout light and dark periods, but was reduced when sleep pressure increased by sleep deprivation at least two hours [31]. Interestingly, the immediate early gene c-Fos, a biomarker of active neurons, was highly expressed in Hcrt neurons regardless of sleep deprivation, but its expression in the downstream arousal-promoting LC and TMN following light activation of Hcrt neurons was less robust with accumulation of sleep pressure [31]. Hcrt neurons of the LH need such downstream neurons, especially noradrenergic neurons in LC, for their wake-promoting function [32]. On the other hand, the optogenetic silencing of Hcrt neurons results in NREM sleep-to-wake transition, which proves that Hcrt neurons are necessary for the maintenance of wakefulness [33]. Hcrt neuropeptides are essential to maintain stable wakefulness [34], and the co-transmission of Hcrt and glutamate have been found to have key implications in the arousal systems [35].
LC noradrenergic neurons are the second subset of neurons involved in sleep-wake circuitry investigated with optogenetics [29]. Light stimulation of LC noradrenergic neurons produced an immediate transition non-REM and REM sleep to wakefulness [36]. A major difference between LC noradrenergic neurons and Hcrt neurons is that optogenetic stimulation of Hcrt cells awaken a sleeping mouse in approximately 30 seconds, whereas stimulating LC neurons require less than five seconds [29]. Tonic but not phasic photoactivation of LC neurons during wake state increased the time spent awake as well as its general locomotor arousal.
The BF an important mediating center of wakefulness also has been investigated with optogenetics. Cholinergic projections from BF neurons modulate the activities of cortical neurons that express neuronal nitric oxide synthase and the receptor for Substance P (NKIR) potentially regulating cortical activity along states of arousal [37]. Although cholinergic neurons are known as wake-promoting neurons [25], the wake-promoting effect of cholinergic neurons is nullified by the pharmacological blockade of local cholinergic receptors in the BF [24]. BF PV neurons have been shown to regulate and entrain cortical gamma band oscillations, which has enhanced the scientific understanding of the mechanisms behind arousal [38].
Recently GABAergic neurons of the bed nucleus of the stria terminalis can induce sudden wakefulness without activating the Hcrt system [39]. Fujita et al. [40] also showed that optogenetic inhibition of HA neurons in the ventral TMN during periods of wakefulness promotes slow wave sleep, but not REM sleep.
SLEEP-RELATED NEURONS DISCOVERED WITH OPTOGENETICSThere have been seminal studies on some neuronal groups which are sleep-active and/or sleep-promoting. Lesions in the preoptic area (POA) can have significant negative consequences on sleep [41]. This suggests that regions of the POA may be linked to the generation of sleep. The VLPO is the most well-known area among sleep-active and sleep-promoting areas, including the medial preoptic area, BF and LH [17,42]. Projections from the VLPO release GABA and galanin onto the wake-related nuclei. More specifically, bidirectional optogenetic manipulation revealed that GABAergic neurons in the preoptic area projecting to the TMN are sleep promoting as well as sleep active [43].
There have been a number of studies on NREM sleep. Sleep spindles have long been used as an indicator of stage 2 of NREM sleep. However, the direct causal relationship between sleep spindles and the stability of the sleep had been questioned. In one study, optogenetic stimulation of the thalamic reticular nucleus was found to increase sleep spindles, the density of which are positively correlated with the amount of NREM sleep [44]. Emotional factors also have an effect on sleep-wake behavior. For example, in the NAc, adenosine plays a role influencing behavioral arousal. Optogenetic activation of core NAc adenosine A2A receptor-expressing (A2AR) neurons greatly promotes slow wave sleep (SWS), demonstrating a noteworthy association of a sleep control being with motivational behavior [45].
In the brain stem, activation of GABAergic cells of the medullary parafacial zone induces SWS by inhibiting parabrachial nucleus which sends the excitatory signal to the BF [46]. In NREM sleep, neuronal oscillations occur between the cortical and thalamic neurons. With the cortical neurons, the off-states of these cells occur nearly simultaneously with one another. A study combining optogenetic and chemogenetic techniques investigated somatostatin-positive (SOM) inhibitory interneurons in the cortex revealed a connection between cortical SOM neurons and the generation of the slow waves in NREM sleep [47].
On the other hand, some neuronal groups are related to REM sleep. REM sleep is believed to be regulated, at least in part, by cholinergic neurons of the mesopontine tegmentum, which are known as “REM ON cells.” However, lesions in this area have not had consistent effects on REM sleep. Investigations utilizing optogenetic activation to determine the role of cholinergic neurons of the PPT or LDT during NREM sleep resulted in an increase in REM sleep episodes without altering REM sleep episode duration [48].
Melanin concentrating hormone (MCH) neurons have also been considered as REM sleep-related neurons [49–52], whereas their function on NREM sleep is still controversial [51–53]. Whereas Konadhode et al. [50] showed increased duration of both NREM and REM sleep, Jego et al. [52] only that of REM sleep. Jego et al. [52] demonstrated that GABA is released as a result of optogenetic stimulation of MCH neurons’ axon terminals projected to the TMN with in vitro recording of the brain slice and their in vivo optogenetic stimulation resulted in increased duration of REM sleep [52]. Additionally, MCH neurons play an essential role on generating theta rhythms during REM sleep, which is necessary for contextual memory consolidation, via projection to the medial septum [49,53].
REM sleep is often affected by mood, stress and fear, but their relationships are complex [54]. It has been hypothesized that REM sleep plays a role in the emotional management of traumatic experiences [54,55]. Machida et al. [54] demonstrated that stress-induced reduction of REM sleep was attenuated by inhibiting glutamatergic neurons of the basolateral nuclei of the amygdala (BLA). This finding suggests that glutamatergic neurons in BLA may interfere the normal physiology of REM sleep generation in response to acute stress.
SLEEP-RELATED MEMORY CONSOLIDATION AND NEUROPLASTICITYWhether or not there is an association between sleep and memory consolidation has been debated for almost a century and there is still no consensus [56,57]. In 1983, it had been proposed that REM sleep might function to unlearn unneeded memory traces and prevent the formation of parasitic modes [58]. Between 1995 and 2005, research on the relationship between sleep and memory increased five-fold, with the majority supporting the role of sleep in “offline” processing of memory [59]. Much of the evidence in support of the REM sleep memory consolidation hypothesis was based around three main evidences. The first was that learning resulted in increased durations in REM sleep [60]. The second evidence was that memory processing occurred during REM sleep [60]. And the third evidence was that the deprivation of REM sleep resulted in the impaired memory consolidation [60]. A separate study conducted by Boyce et al. [53] identified a connection between GABAergic neurons of the medial septum (MS) and memory consolidation during REM sleep by demonstrating that optogenetic silencing of MSGABA neurons impaired novel object recognition abilities of mice. Furthermore, a study by Rolls et al. [57] demonstrated that sleep fragmentation impairs memory consolidation. Although studies have determined that memories in pattern recognition or procedures improve after sleeping, memorization of facts do not appear to improve after sleep [56]. Thus, many researchers believe that the central hypothesis suggesting that information acquired throughout a day is strengthened during sleep still needs further evidence [56]. In 2011, a group of researchers optogenetically stimulated Hcrt neurons to cause sleep fragmentation without effecting total sleep duration, sleep quality, or intensity, resulting in impaired memory consolidation [57].
NREM sleep has also been identified as essential for the memory consolidation of motor and sense based learning experiences [61]. Using optogenetics, Miyamoto et al. [62] determined that perceptual memory is consolidated during NREM sleep via top-down cortical input. Optogenetic inactivation of top-down projecting secondary motor cortex (M2) neurons to primary somatosensory cortex (S1) neurons impaired memory consolidation and slow wave synchrony in the cortex is necessary for the top-down cortical control between M2 and S1 [62]. Research by Latchoumane et al. [63] found that hippocampus-dependent memory can be disrupted by optogenetic suppression of thalamic sleep spindles of NREM sleep suggesting a role for the sleep spindles in memory consolidation. They suggested that triple coupling of slow oscillations, spindles, and hippocampal ripples are essential for the memory.
Sleep also has a significant influence on neuroplasticity. Durkin et al. [64] investigated the potential function of NREM thalamocortical oscillations in neuroplasticity. Their study concluded that NREM oscillations, coordinated by corticothalamic communication from visual cortex to lateral geniculate nucleus, play a role in relaying information of previously collected sensory experiences, thereby promoting cortical plasticity [64].
OPTOGENETIC STUDIES RELATED TO SLEEP DISORDERSObstructive Sleep ApneaObstructive sleep apnea is one of the major sleep disorders involving serious health consequences such as cardiovascular events. During apnea episodes, hypoxia and hypercapnia, two major adverse physiological changes, occur simultaneously. Recently these two phenomena were investigated separately, using optogenetics. C1 neurons in the rostral ventrolateral medulla are believed to regulate blood pressure as well as stimulate nuclei in the central nervous system involved in breathing, vigilance and stress responses [65]. Burke et al. [66] optogenetically activated C1 neurons during NREM sleep and found EEG de-synchronization and cardiorespiratory stimulation. They suggested that C1 cells, specifically during NREM sleep, may exert a critical influence on the sleep disruption and adverse autonomic consequences of sleep apnea. C1 cell activation reproduced similar physiological effects to those observed in hypoxia, which supports the idea that C1 cells are at least partially responsible for the cardiovascular responses and arousal from sleep during apnea events [66]. C1 cells potentially generate arousal through a variety of mechanisms including activation of the LC, raphe, orexinergic neurons [67,68] or other pathways that are inhibited during REM sleep [69,70].
On the other hand, external lateral parabrachial nucleus (PBel) was investigated to understand the adverse effects of hypercapnia. Especially, when the PBel neurons that express calcitonin gene-related peptide, were stimulated optogenetically and chemogenetically, wakefulness was evidently caused [71]. Conversely, when those neurons were inhibited with ArchT, the arousal by hypercapnia was prevented with a four-fold increase in arousal latency, but not by sound or shaking [71].
NarcolepsyNarcolepsy is caused by a significant loss of Hcrt neurons and/or diminished Hcrt signaling in the brain [72]. As previously mentioned, Adamantidis et al. [30] showed photostimulation of Hcrt neurons in the LH induced wakefulness, which supports that the function of Hcrt is sufficient for causing wakefulness. Their findings substantiated prior findings linking narcolepsy with cataplexy and low counts, often a reduction of between 85% to 95%, of Hcrt neurons in the LH and low levels of Hcrt in the cerebrospinal fluid [73,74]. Results of the loss-of-function studies indicate that narcolepsy with cataplexy is a disorder based on the selective degeneration of Hcrt cells [33,74].
A sudden weakening of muscle tone, or cataplexy, is one of the cardinal symptoms of narcolepsy and serotonergic medication is effective to reduce cataplexy. Using optogenetics, it was shown that cataplexy could be suppressed by inhibition of specific projections of serotonergic neurons of DRN to amygdala [75]. Based on this finding, it was suggested that orexin/Hcrt neurons stimulate DRN serotonin neurons which, in turn, inhibit amygdala in physiological condition. However, in narcolepsy, the lack of orexin/Hcrt resulted in the suppressed activity of DRN and subsequent overactivity of the amygdala as well as the generation of cataplexy.
CONCLUSIONSince optogenetics, one of the state-of-the-art technologies, has been introduced in the field of sleep research, various cell types in different regions of the brain were explored and their functions in sleep-wake control were elucidated. This novel technology opened a flourishing era of sleep research both in validation of pre-existing hypotheses and discovery of new neuronal types and circuitries. Although optogenetics is a strong tool for investigating brain circuitry, it should be noted that the brain is an electrochemical organ which is operated by not only electrical connections but also chemical components like neurotransmitters and neuromodulators. Both the circuitry and neurochemical dynamics should be considered simultaneously. Better understandings of neural networks and signaling pathways will allow researchers to unfold the pathophysiology of sleep disorders and some relevant psychiatric disorders, and to develop novel therapeutic modalities by manipulating sleep circuitry and neurochemical regulations in the near future.
AcknowledgmentsThis research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF 2015R1D1A1A01059119), the Ministry of Science, ICT & Future Planning (NRF 2018R1A2B6002804), the Engineering Research Center of Excellence (ERC) Program (NRF 2017R1A5A1014708), GIST Research Institute (GRI) grant 2018.
NOTESAuthors’ Contribution
Drew VJ performed review conception and design, collection of references, analysis and interpretation of literature, drafting of manuscript. Lee JM performed study conception and design, figure generation, analysis and interpretation of literature. Kim T performed review conception and design, analysis and interpretation of literature, drafting of manuscript, and critical revision.
REFERENCES2. Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep state switching. Neuron 2010;68:1023-42.
3. Scammell TE, Arrigoni E, Lipton JO. Neural circuitry of wakefulness and sleep. Neuron 2017;93:747-65.
4. Van Dongen HP, Maislin G, Mullington JM, Dinges DF. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep 2003;26:117-26.
5. Hillman DR, Lack LC. Public health implications of sleep loss: the community burden. Med J Aust 2013;199:S7-10.
6. de Lecea L, Carter ME, Adamantidis A. Shining light on wakefulness and arousal. Biol Psychiatry 2012;71:1046-52.
7. Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, et al. The microbial opsin family of optogenetic tools. Cell 2011;147:1446-57.
8. Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 2005;8:1263-8.
9. Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci 2009;12:229-34.
10. Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O’Shea DJ, Prakash R, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods 2011;9:159-72.
11. Guru A, Post RJ, Ho YY, Warden MR. Making sense of optogenetics. Int J Neuropsychopharmacol 2015;18:pyv079.
12. Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc 2010;5:439-56.
13. Luo L, Callaway EM, Svoboda K. Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 2018;98:256-81.
14. Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, et al. Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of Channelrhodopsin-2. Nat Protoc 2010;5:247-54.
15. Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci 2012;15:793-802.
16. Jones BE. Neurobiology of waking and sleeping. In Montagna P, Chokroverty S (eds.), Handb Clin Neurol. 98:Amsterdam: Elsevier; 2011. pp. 131-49.
17. Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature 2005;437:1257-63.
18. Gaus SE, Strecker RE, Tate BA, Parker RA, Saper CB. Ventrolateral preoptic nucleus contains sleep-active, galaninergic neurons in multiple mammalian species. Neuroscience 2002;115:285-94.
19. Sherin JE, Elmquist JK, Torrealba F, Saper CB. Innervation of histaminergic tuberomammillary neurons by GABAergic and galaninergic neurons in the ventrolateral preoptic nucleus of the rat. J Neurosci 1998;18:4705-21.
20. Gallopin T, Fort P, Eggermann E, Cauli B, Luppi PH, Rossier J, et al. Identification of sleep-promoting neurons in vitro. Nature 2000;404:992-5.
21. Ito H, Yanase M, Yamashita A, Kitabatake C, Hamada A, Suhara Y, et al. Analysis of sleep disorders under pain using an optogenetic tool: possible involvement of the activation of dorsal raphe nucleus-serotonergic neurons. Mol Brain 2013;6:59.
22. Cho JR, Treweek JB, Robinson JE, Xiao C, Bremner LR, Greenbaum A, et al. Dorsal Raphe Dopamine Neurons Modulate Arousal and Promote Wakefulness by Salient Stimuli. Neuron 2017;94:1205-19.e8.
23. Yu X, Ye Z, Houston CM, Zecharia AY, Ma Y, Zhang Z, et al. Wakefulness Is Governed by GABA and Histamine Cotransmission. Neuron 2015;87:164-78.
24. Zant JC, Kim T, Prokai L, Szarka S, McNally J, McKenna JT, et al. Cholinergic neurons in the basal forebrain promote wakefulness by actions on neighboring non-cholinergic neurons: an opto-dialysis study. J Neurosci 2016;36:2057-67.
25. Xu M, Chung S, Zhang S, Zhong P, Ma C, Chang WC, et al. Basal forebrain circuit for sleep-wake control. Nat Neurosci 2015;18:1641-7.
26. Ozen Irmak S, de Lecea L. Basal forebrain cholinergic modulation of sleep transitions. Sleep 2014;37:1941-51.
27. Eban-Rothschild A, Rothschild G, Giardino WJ, Jones JR, de Lecea L. VTA dopaminergic neurons regulate ethologically relevant sleep-wake behaviors. Nat Neurosci 2016;19:1356-66.
28. De Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE, et al. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci USA 1998;95:322-7.
29. Touriño C, Eban-Rothschild A, de Lecea L. Optogenetics in psychiatric diseases. Curr Opin Neurobiol 2013;23:430-5.
30. Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 2007;450:420-4.
31. Carter ME, Adamantidis A, Ohtsu H, Deisseroth K, de Lecea L. Sleep homeostasis modulates hypocretin-mediated sleep-to-wake transitions. J Neurosci 2009;29:10939-49.
32. Carter ME, Brill J, Bonnavion P, Huguenard JR, Huerta R, de Lecea L. Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc Natl Acad Sci USA 2012;109:E2635-44.
33. Tsunematsu T, Kilduff TS, Boyden ES, Takahashi S, Tominaga M, Yamanaka A. Acute optogenetic silencing of orexin/hypocretin neurons induces slow-wave sleep in mice. J Neurosci 2011;31:10529-39.
34. Liblau RS, Vassalli A, Seifinejad A, Tafti M. Hypocretin (orexin) biology and the pathophysiology of narcolepsy with cataplexy. Lancet Neurol 2015;14:318-28.
35. Schöne C, Apergis-Schoute J, Sakurai T, Adamantidis A, Burdakov D. Coreleased orexin and glutamate evoke nonredundant spike outputs and computations in histamine neurons. Cell Rep 2014;7:697-704.
36. Carter ME, Yizhar O, Chikahisa S, Nguyen H, Adamantidis A, Nishino S, et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat Neurosci 2010;13:1526-33.
37. Williams RH, Vazquez-DeRose J, Thomas AM, Piquet J, Cauli B, Kilduff TS. Cortical nNOS/NK1 Receptor Neurons are Regulated by Cholinergic Projections From the Basal Forebrain. Cereb Cortex 2018;28:1959-79.
38. Kim T, Thankachan S, McKenna JT, McNally JM, Yang C, Choi JH, et al. Cortically projecting basal forebrain parvalbumin neurons regulate cortical gamma band oscillations. Proc Natl Acad Sci USA 2015;112:3535-40.
39. Kodani S, Soya S, Sakurai T. Excitation of GABAergic neurons in the bed nucleus of the stria terminalis triggers immediate transition from non-rapid eye movement sleep to wakefulness in mice. J Neurosci 2017;37:7164-76.
40. Fujita A, Bonnavion P, Wilson MH, Mickelsen LE, Bloit J, de Lecea L, et al. Hypothalamic tuberomammillary nucleus neurons: electrophysiological diversity and essential role in arousal stability. J Neurosci 2017;37:9574-92.
41. Sallanon M, Denoyer M, Kitahama K, Aubert C, Gay N, Jouvet M. Long-lasting insomnia induced by preoptic neuron lesions and its transient reversal by muscimol injection into the posterior hypothalamus in the cat. Neuroscience 1989;32:669-83.
42. Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW. Control of sleep and wakefulness. Physiol Rev 2012;92:1087-187.
43. Chung S, Weber F, Zhong P, Tan CL, Nguyen TN, Beier KT, et al. Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 2017;545:477-81.
44. Kim A, Latchoumane C, Lee S, Kim GB, Cheong E, Augustine GJ, et al. Optogenetically induced sleep spindle rhythms alter sleep architectures in mice. Proc Natl Acad Sci USA 2012;109:20673-8.
45. Oishi Y, Xu Q, Wang L, Zhang BJ, Takahashi K, Takata Y, et al. Slow-wave sleep is controlled by a subset of nucleus accumbens core neurons in mice. Nat Commun 2017;8:734.
46. Anaclet C, Ferrari L, Arrigoni E, Bass CE, Saper CB, Lu J, et al. The GABAergic parafacial zone is a medullary slow wave sleep-promoting center. Nat Neurosci 2014;17:1217-24.
47. Funk CM, Peelman K, Bellesi M, Marshall W, Cirelli C, Tononi G. Role of somatostatin-positive cortical interneurons in the generation of sleep slow waves. J Neurosci 2017;37:9132-48.
48. Van Dort CJ, Zachs DP, Kenny JD, Zheng S, Goldblum RR, Gelwan NA, et al. Optogenetic activation of cholinergic neurons in the PPT or LDT induces REM sleep. Proc Natl Acad Sci USA 2015;112:584-9.
49. Luppi PH, Peyron C, Fort P. Role of MCH neurons in paradoxical (REM) sleep control. Sleep 2013;36:1775-6.
50. Konadhode RR, Pelluru D, Blanco-Centurion C, Zayachkivsky A, Liu M, Uhde T, et al. Optogenetic stimulation of MCH neurons increases sleep. J Neurosci 2013;33:10257-63.
51. Hassani OK, Lee MG, Jones BE. Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep-wake cycle. Proc Natl Acad Sci USA 2009;106:2418-22.
52. Jego S, Glasgow SD, Herrera CG, Ekstrand M, Reed SJ, Boyce R, et al. Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat Neurosci 2013;16:1637-43.
53. Boyce R, Glasgow SD, Williams S, Adamantidis A. Causal evidence for the role of REM sleep theta rhythm in contextual memory consolidation. Science 2016;352:812-6.
54. Machida M, Wellman LL, Fitzpatrick Bs ME, Hallum Bs O, Sutton Bs AM, Lonart G, et al. Effects of optogenetic inhibition of BLA on sleep brief optogenetic inhibition of the basolateral amygdala in mice alters effects of stressful experiences on rapid eye movement sleep. Sleep 2017;40:zsx020.
55. van der Helm E, Yao J, Dutt S, Rao V, Saletin JM, Walker MP. REM sleep depotentiates amygdala activity to previous emotional experiences. Curr Biol 2011;21:2029-32.
56. Higgins ES, George MS. Neuroscience of clinical psychiatry: the pathophysiology of behavior and mental illness. 2nd ed. Philadelphia: Lippincott Williams & Wilkins; 2013.
57. Rolls A, Colas D, Adamantidis A, Carter M, Lanre-Amos T, Heller HC, et al. Optogenetic disruption of sleep continuity impairs memory consolidation. Proc Natl Acad Sci USA 2011;108:13305-10.
61. Yang G, Lai CS, Cichon J, Ma L, Li W, Gan WB. Sleep promotes branch-specific formation of dendritic spines after learning. Science 2014;344:1173-8.
62. Miyamoto D, Hirai D, Fung CC, Inutsuka A, Odagawa M, Suzuki T, et al. Top-down cortical input during NREM sleep consolidates perceptual memory. Science 2016;352:1315-8.
63. Latchoumane CV, Ngo HV, Born J, Shin HS. Thalamic spindles promote memory formation during sleep through triple phase-locking of cortical, thalamic, and hippocampal rhythms. Neuron 2017;95:424-35.e6.
64. Durkin J, Suresh AK, Colbath J, Broussard C, Wu J, Zochowski M, et al. Cortically coordinated NREM thalamocortical oscillations play an essential, instructive role in visual system plasticity. Proc Natl Acad Sci USA 2017;114:10485-90.
65. Ayas NT, Owens RL, Kheirandish-Gozal L. Update in sleep medicine 2014. Am J Respir Crit Care Med 2015;192:415-20.
66. Burke PG, Abbott SB, Coates MB, Viar KE, Stornetta RL, Guyenet PG. Optogenetic stimulation of adrenergic C1 neurons causes sleep state-dependent cardiorespiratory stimulation and arousal with sighs in rats. Am J Respir Crit Care Med 2014;190:1301-10.
67. Holloway BB, Stornetta RL, Bochorishvili G, Erisir A, Viar KE, Guyenet PG. Monosynaptic glutamatergic activation of locus coeruleus and other lower brainstem noradrenergic neurons by the C1 cells in mice. J Neurosci 2013;33:18792-805.
68. Guyenet PG, Abbott SB. Chemoreception and asphyxia-induced arousal. Respir Physiol Neurobiol 2013;188:333-43.
69. Aston-Jones G, Bloom FE. Activity of norepinephrine-containing locus coeruleus neurons in behaving rats anticipates fluctuations in the sleep-waking cycle. J Neurosci 1981;1:876-86.
70. Lee MG, Hassani OK, Jones BE. Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle. J Neurosci 2005;25:6716-20.
71. Kaur S, Wang JL, Ferrari L, Thankachan S, Kroeger D, Venner A, et al. A genetically defined circuit for arousal from sleep during hypercapnia. Neuron 2017;96:1153-67.e5.
73. Thannickal TC, Moore RY, Nienhuis R, Ramanathan L, Gulyani S, Aldrich M, et al. Reduced number of hypocretin neurons in human narcolepsy. Neuron 2000;27:469-74.
Fig. 1Optogenetics and sleep research. A: Schematic diagram of optogenetics. Optogenetics is characterized with the utilization of opsins, light-sensitive proteins, and light of specific wavelengths to excite or inhibit specific neuronal types. For example, ChR2 expressed in the neurons can cause action potentials by causing Na+ influx when illuminated by blue light, whereas NpHR can inhibit action potentials by causing Cl− influx when illuminated by yellow light. An opsin gene can be packaged into viral vector, which is injected into the region of interest stereotactically. B: Application of optogenetics in sleep research. Wake- and sleep-related neural circuitries have been investigated using optogenetics. Blue and red circles represent wake- and sleep-related regions in the brain. BF: basal forebrain, BNST: bed nucleus of the stria terminalis, ChR2: channelrhodopsin-2, DRN: dorsal raphe nucleus, LC: locus coeruleus, LH: lateral hypothalamus, M2: secondary motor cortex, NACC: nucleus accumbens core, nm: nanometer, NpHR: halorhodopsin, POA: pre-optic area, PPT: pedunculopontine tegmental nucleus, PZ: parafacial zone, VTA: ventral tegmental area. 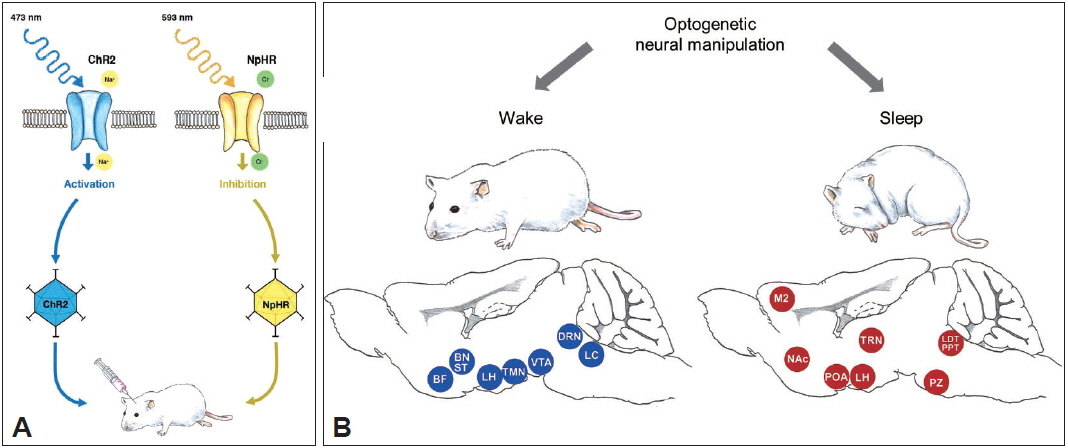
Table 1Summarization of recent publications on the sleep-wake control using optogenetics
A2AR: adenosine A2A receptor, AAV: adeno-associated virus, ACh: acetylcholine, ArchT: archaerhodopsin from halorubrum strain TP009, BF: basal forebrain, BNST: bed nucleus of the stria terminalis, C1: catecholaminergic neurons, C1V1: chimeric opsin of ChR1 and volvox ChR1, CaMKIIa: calcium/calmodulin-dependent protein kinase II alpha, CGRP: calcitonin generelated peptide, ChR2: channelrhodopsin 2, ChAT: choline acetyltransferase, ChETA: ChR2 variant replacing active site residue E123 by Thr and Ala, CT: corticothalamic neurons, DA: dopaminergic, DIO: double floxed-inverse open reading frame, DRN: dorsal raphe nuclei, eNpHR: enhanced halorhodopsin, eGFP: enhanced green fluorescence protein, eYFP: enhanced yellow fluorescence protein, FLEX: floxed-inversed, GAD67: glutamic acid decarboxylase 67, Glu: glutamatergic, HA: histaminergic, Hcrt: hypocretin/orexin, Hdc: histidine decarboxylase, 5-HT: serotonergic, LC: locus coeruleus, LDT: laterodorsal tegmental nucleus, LH: lateral hypothalamus, M2: secondary motor cortex, MCH: melanin-concentrating hormone, MS: medial septum, NAc: nucleus accumbens, NE: noradrenergic, NK1R: neurokinin 1 receptor, nNOS: neuronal nitric oxide synthase, Ntsr1: neurotepsin receptor 1, PB: parabrachial nucleus, Pbel: external parabrachial nucleus, PPT: pedunculopontine tegmental nucleus, PV: parvalbumin, PZ: parafacial zone, RVLM: rostral ventrolateral medulla, SOM: somatostatin, SSFO: stabilized step-function opsin, SuM: supramammillary nucleus, SWS: slow wave sleep, TH: tyrosine hydroxylase, TMN: tuberomammillary nucleus, TRN: thalamic reticular nucleus, Vgat: vesicular GABA transporter, Vglut2: vesicular glutamate transporter 2, VTA: ventral tegmental area. |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||